Is it Possible that DNA Genetic Material itself to fool the Definite Determination? Part VI - a recap and expansion
Real Fathers may escape responsibility of fathering; Lawyers shall stand on their toes on floor to question the protocols used by laboratories, to determine a real father.
In the last episode we saw that there existed
many hurdles in using DNA typing for any forensic investigation. The amount of
sample that have to be collected, and adequate amount of DNA that can be
extracted for the analytical methods in use during nineteen eighties and into
the early nineteen nineties were of very challenging. Extraction of DNA became sensationally
possible virtually from any tiny material and analytical procedures as well
improved day by day, that made the required amount DNA for a procedure also,
very minimal with even more sensationally. Having solved these, the second
hurdle faced by scientists was target sequences within the DNA extracted.
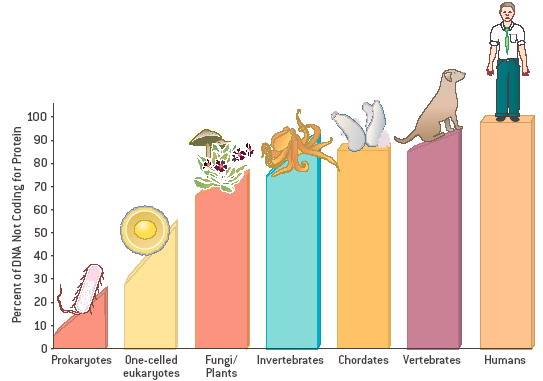
Originally they were targeting coding sequences of real genes which is only
represents a minimal 2% of entire Nucleotide base pairs of 3.3 billion for a
human and other mammals and vertebrates bit higher 3% to 20%. This gene codon
targets gave enough trouble to laboratories since mutations in DNA, due to
environmental pressure and impacts. Human usually lives with about 400 odd
Mutations within DNA without any apparent defects. It is best to remember loci length are
measured in base pairs (bp) and we saw that using intron became ethical,
the obvious reasons for that the noncoding intronic codons are very
conservative do not easily undergo mutation during the replication of DNA, higher the animal in evolution tree, higher the
percentage of noncoding introns within the DNA helix molecule. Repetitive
sequences between two loci very distinctive, and numerous making it easy
specific for targets for forensic and
anthropological studies.
Once the targets have been located
within the extracted DNA, fragmenting is a relatively easy process by the use
of specific enzymes as it acts only on targets.
Mitochondrial DNA does not contain Non-coding Introns. Anthropological
studies and Forensic science markedly improved mainly by ability to recover DNA
that can be processed from frozen and dormant biological as well as from inert
materials, and main the objective of polymerase chain
reaction (PCR) is to produce enough DNA
template and huge number of copies of each to work with for further
analysis and typing from recovered sample.
Polymerase Chain
Reaction (PCR)
A revolutionary
method developed by Kary Mullis in the 1980s. PCR is based on using the ability
of DNA polymerase to
synthesize new strand of DNA complementary to the offered template strand. The
first and most commonly used of these enzymes is Taq DNA
polymerase (from bacteria Thermis aquaticus), whereas Pfu DNA
polymerase (from Bacteria Pyrococcus furiosus) is used widely because of its
higher accuracy when copying DNA. Although these enzymes are subtly different,
they both have two capabilities that make them suitable for PCR because they can
generate new strands of DNA using a DNA template and primers - single units of the bases A, T, G, and C, which are essentially "building blocks" for new DNA strands and because essentally they are
heat resistant, Extremely variable regions, characteristic of non-coding DNA are used.
Genetically different individuals produce different profiles. The closer the
genetic relationship between individuals the more similar their profiles
1. The DNA is released from a cell: The
cells of an uncontaminated biological sample (blood, semen, hair root, cheek
cell) are broken open and the DNA is released and separated. The method relies on thermal cycling,
consisting of cycles of repeated heating and cooling of the reaction within in a tube for DNA melting and enzymatic replication of the DNA. Primers (short DNA fragments) containing sequences complementary to
the target region along with a DNA polymerase
Because DNA polymerase
can add a nucleotide only onto a preexisting 3'-OH group, it needs a primer to which it can add the first nucleotide. This requirement
makes it possible to delineate a specific region of template sequence that the
researcher wants to amplify. At the end of the PCR reaction, the specific
sequence will be accumulated in billions of copies (amplicons).
2.
There are 3 steps
initially
A.
The cycling reactions :
The three initial steps
in a PCR that are repeated for 30 or 40 cycles. This is done on an automated machine which can heat and cool the tubes with the reaction mixture in a very
short time. I. Denaturation at
94°C: during the denaturation, the double strand melts open to single stranded
DNA, all enzymatic reactions stop (for example : the extension from a previous
cycle) II. Annealing at
54°C: the primers are jiggling around, caused by the Brownian motion. Ionic
bonds are constantly formed and broken between the single stranded primer and
the single stranded template. The more stable bonds last a little bit longer
(primers that fit exactly) and on that little piece of double stranded
DNA (template and primer), the polymerase can attach and starts copying
the template. Once there are a few bases built in, the ionic bond is so
strong between the template and the primer, that it does not break anymore. III. Extension at
72°C: This is the ideal working temperature for the polymerase. The
primers, where there are a few bases built in, already have a stronger ionic
attraction to the template than the forces breaking these attractions. Primers
that are on positions with no exact match, get loose again (because of the
higher temperature) and don't give an extension of fragment. The bases
(complementary to the template) are coupled to the primer on the 3' side (the
polymerase adds dNTP's from 5' to 3', reading the template from 3' to 5' side,
bases are added complementary to the template)
B.
Is
there a gene copied during PCR and is it the right size?
Before the PCR product is used in further
applications, it has to be checked if
a.
There is a product
formed. Though biochemistry is an exact science, not every PCR is
successful. There is for example a possibility that the quality of the DNA is
poor, that one of the primers doesn't fit, or that there is too much starting
template
b.
The product is of the
right size, It is possible that there is a product, for example a band of 500
bases, but the expected gene should be 1800 bases long. In that case, one of
the primers probably fits on a part of the gene closer to the other primer. It
is also possible that both primers fit on a totally different gene.
c. Only one band is formed. As in the description
above, it is possible that the primers fit on the desired locations, and also
on other locations. In that case, you can have different bands in one lane on a
gel.
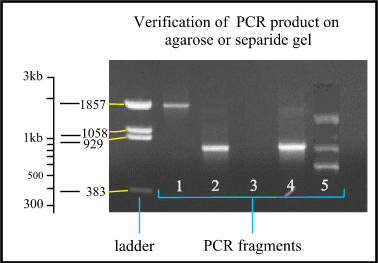
Electrophoresis
Verification,
The ladder is a mixture
of fragments with known size to compare with the PCR fragments. Notice that the
distance between the different fragments of the ladder is logarithmic. Lane 1:
PCR fragment is approximately 1850 bases long. Lane 2 and 4: the fragments are
approximately 800 bases long. Lane 3: no product is formed, so the PCR failed.
Lane 5: multiple bands are formed because one of the primers fits on different
places. a constrain of PCR method.
Having completed enough
DNA recovering process, actual analysis commenced as described last
episode.
3. Digestion: Special enzymes are used to cut the DNA at specific points and produce a set of
fragments of varying lengths. These enzymes are called restriction enzymes.
The DNA sections that are cut are called restriction fragments. Certain enzymes will cut the DNA at specific bases (T, G,
A, and C). Each restrictive fragment will be specific for that particular
person. The bases as well as the distance between the bases (non-coding DNA) are characteristics
of that individual organism (person).
4. Separation: The fragment mixture is
placed in a block of gel and separated by gel electrophoresis on the basis of size. The fragments are exposed to an electrical
charge. This causes the fragments to move. The smaller the fragment the further
it travels. The bands of small fragments are separated from the bands of the
large fragments. After other treatments, a photographic copy of the pattern of
DNA is compiled. Using trade mark method the positions of the DNA fragments on
X-ray film appear as dark bands. The positions of these bands are the genetic
profile of the individual and the more numerous the bands the more reliable the
‘fingerprint’. The bands are produced and vary in thickness depending on how
many DNA fragments are present in a certain length of DNA.
5. Pattern comparison: The DNA fingerprint can be used to be compared to other DNA
fingerprints. The pattern is unique to an individual. In this way comparisons
of known DNA to unknown DNA can be made to determine if the unknown DNA can be
identified.
Applications
of DNA Profiling
- Crime:
Forensic medicine uses medical evidence in legal
disputes. DNA profiling is used in forensic medicine.
Material such as hair, semen, blood, and saliva which is left at a crime
scene can be compared with possible suspects to find out if the suspect
was at the crime scene.
- Medical: DNA profiles can be used to determine the parents of a child.
GENETIC SCREENING
Genetic Screening is used to find out if the parents of a
child or the child itself (through removal of cells from the foetus) carry
defective genes that could develop into health problems. Although the parent
may not have the disorder he/she could be a carrier for the condition. With genetic screening, prospective
parents can determine if there is a possibility that
their future children could have the health problem.



No comments:
Post a Comment